Large-scale genome-wide studies have revealed that only 2% of the human genome encodes for mRNAs that are translated into proteins while as much as 80% of the genome can be transcribed. Of these non-coding RNA transcripts, long non-coding RNAs (lncRNAs) represent the largest and most diverse class. Genome-wide analyses have identified over 19,000 human lncRNA genes representing approximately 31% of annotated human genes. As only a relatively small number of these lncRNAs have been functionally characterized and studied in the context of cancer, they represent a unexplored class of nucleic acid regulatory molecules with great potential to impact our insights into cancer biology and treatment. LncRNAs tend to localize to cell nuclei implying roles in gene expression and/or nuclear organization. Our laboratory has played a pivotal role in identifying and characterizing lncRNAs that are involved in breast cancer progression. In addition, we pioneered the use of antisense oligonucleotides (ASOs) to knockdown lncRNAs demonstrating their therapeutic potential.
With breast cancer being the most frequent malignancy in women worldwide, a major focus of our laboratory has been to identify and characterize lncRNAs that play roles in breast cancer progression, and evaluate their mechanism of action and potential as therapeutic targets. Tumor organoids provide a very innovative and unique platform to study cancer, as they can recapitulate many aspects of the disease with high fidelity. As such they represent an excellent system for identifying new therapeutic targets and for drug development and screening in a patient-specific manner. We have developed and characterized a patient-derived breast tumor organoid biobank to identify new therapeutic targets and for drug development and screening in a patient-specific manner. We performed a comprehensive genomic, transcriptomic, and cellular analysis of 87 breast cancer patient-derived organoid (PDO) lines developed in our lab, out of which 50 have been fully validated as being tumor-derived and retain key genomic properties of breast cancers including SNVs and CNAs.
Using our RNA-seq expression data from triple-negative breast cancer (TNBC) and invasive lobular carcinoma of the breast (ILC) vs normal breast organoids, we performed differential gene expression analysis and we identified ~200 lncRNAs that are upregulated in TNBC or ILC PDOs vs normal breast organoids. We cross referenced these lists with lncRNAs found to be upregulated in TCGA breast cancer cases, to identify robust lists of intergenic TNBC and ILC lncRNA candidates that are the focus of our current studies.
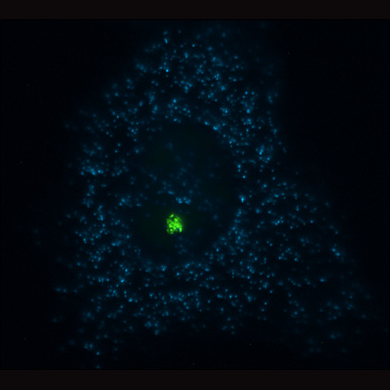
Live cell visualization of a stably integrated genetic locus (yellow/green) using the lac operator/repressor system and yellow fluorescent protein. In the transcriptionally active state the locus is decondensed and transcribes an RNA that encodes for a cyan fluorescent protein fusion that is targeted to cytoplasmic peroxisomes. Photo provided by Susan M. Janicki
Tsukamoto, T., N. Hashiguchi, S.M. Janicki, T. Tumbar, A.S. Belmont and D.L. Spector. 2000. Visualization of gene activity in living cells. Nature Cell Biology 2: 871-878. Download PDF
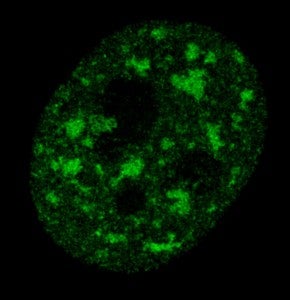
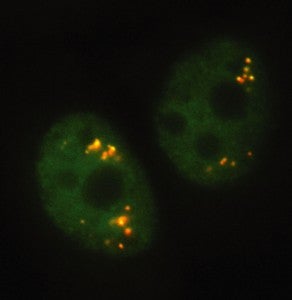
Speckles are subnuclear structures enriched in pre-mRNA splicing factors, located in the interchromatin regions of the nucleoplasm of mammalian cells. At the fluorescence microscope level they appear as irregular, punctate structures that vary in size and shape. When examined by electron microscopy they correspond to clusters of interchromatin granules. Speckles are dynamic structures and both their protein and RNA-protein components can cycle continuously between speckles and other nuclear locations, including active transcription sites.
Mintz, P., S.D. Patterson, A.F. Neuwald, C.S. Spahr, and D.L. Spector. 1999. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 18, 4308-4320.
Lamond, A.I. and D.L. Spector. 2003. Nuclear speckles: a model for nuclear organelles. Nature Reviews: Molec. and Cell. Biol. 4: 605-612. Download PDF
Long non-coding (lncRNAs) are emerging key factors in the regulation of various cellular processes. In the nucleus, these include the organization of nuclear sub-structures, the alteration of chromatin state, and the regulation of gene expression through the interaction with effector proteins and modulation of their activity. Collectively, lncRNAs form the core of attractive models explaining aspects of structural and dynamic regulation in the nucleus across time and space.
Bergmann, J.H. and Spector, D.L. 2014. Long Non-coding RNAs: modulators of nuclear structure and function. Current Opinion in Cell Biol. 26, 10-18. Download PDF